Human DNA Cytosine Methyltransferases
The human DNA cytosine methyltransferase 3A (DNMT3A) is our current focus. This enzyme plays essential roles in early development, and occurs in stem cells in adults, and inappropriately, in cancer cells. DNA methylation at CpG sites controls the expression of proximal genes. Mutations in DNMT3A are some of the most common mutations in diverse cancers. Our interest is to understand how this enzyme is regulated by protein-protein interactions, histone modifications, and non-coding RNA. Our current model shows that non-coding RNA is a dominant form of regulation, although a large number of regulatory proteins as well as histone modifications play important roles as well. Our ultimate goal is to develop small molecules (inhibitors) that don’t show the toxicity of currently used DNMT3A inhibitors. Our recent discovery and design of allosteric inhibitors show exciting promise.
Antibiotic and Cancer Drug Design
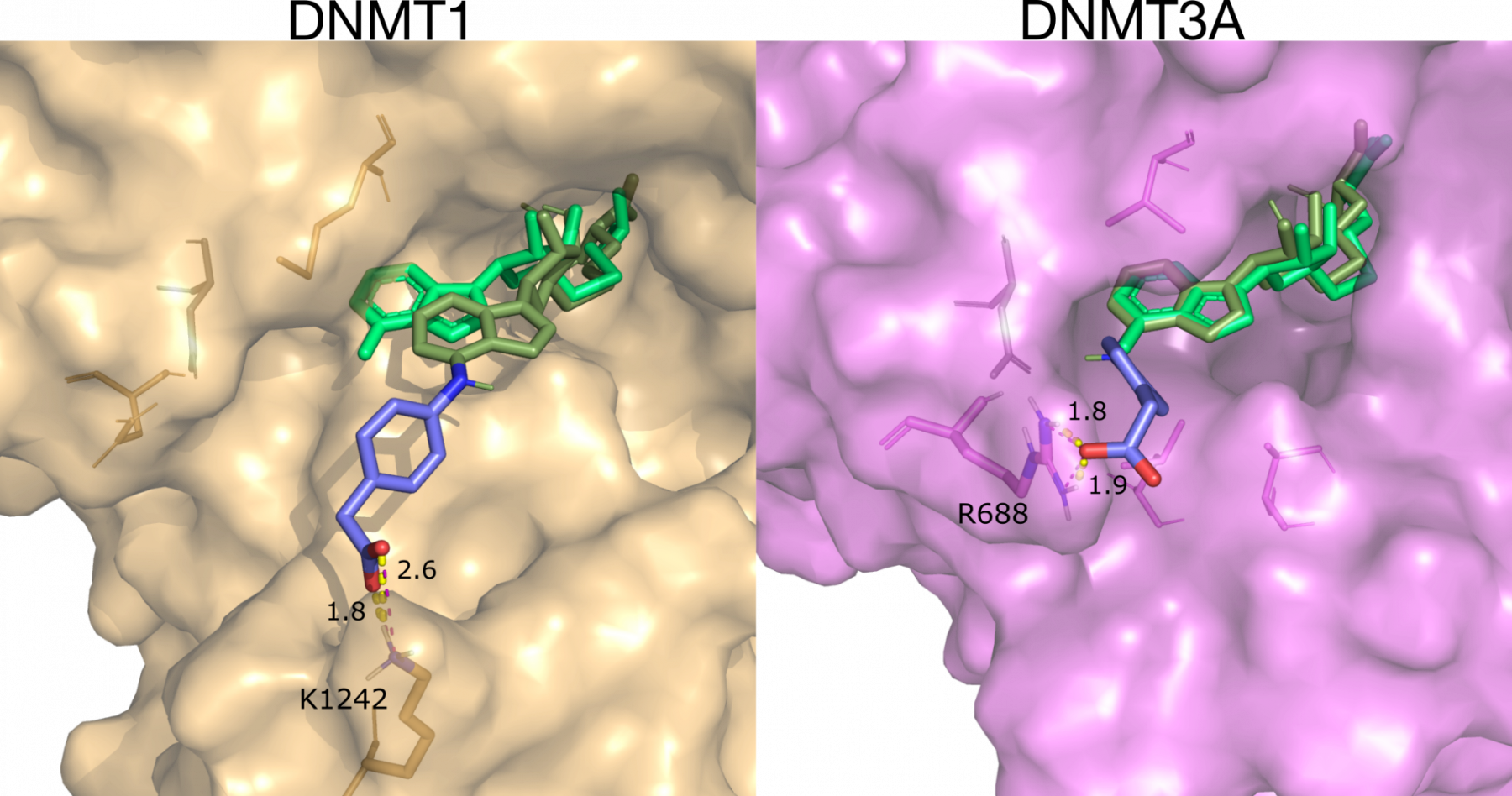
We use our understanding of bacterial and human DNA methyltransferases to design potential antibiotics and cancer treatments, respectively. For example, currently used human DNA methyltransferase (DNMTs) drugs are highly toxic since they show no selectivity for a particular DNMT. Using computational methods, we have designed novel reversible and irreversible inhibitors that selectively target DNMT3A which is implicated in cancer progression (submitted, See Figure, below). We have also discovered allosteric inhibitors of DNMT3A which disrupt protein-protein interactions, and are an attractive and entirely new approach to control epigenetic pathways (submitted).
Related Publications
p53 and TDG are dominant in regulating the activity of the human de novo DNA methyltransferase DNMT3A on nucleosomes
Sandoval JE, Reich NO. J. Biol. Chem., 296 (2021), 100058
https://doi.org/10.1074/jbc.RA120.016125
A novel class of selective non-nucleoside inhibitors of human DNA methyltransferase 3A
Huang S, Stillson NJ, Sandoval JE, Yung C, Reich NO. Bioorg. Med. Chem. Lett., 40 (2021), 127908
https://doi.org/10.1016/j.bmcl.2021.127908
The R882H substitution in the human de novo DNA methyltransferase DNMT3A disrupts allosteric regulation by the tumor supressor p53
Sandoval JE, Reich NO. J. Bio. Chem., 294(48) (2019), 18207-18219
https://doi.org/10.1074/jbc.RA119.010827
Mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia patients cause both loss and gain of function and differential regulation by protein partners
Sandoval JE, Huang YH, Muise A, Goodell MA, Reich NO. J. Bio. Chem., 294(13) (2019), 4898-4910
https://doi.org/10.1074/jbc.RA118.006795
De novo DNA methyltransferase DNMT3A: Regulation of oligomeric state and mechanism of action in response to pH changes
Holz-Schietinger C, Reich NO. BBA-Gen. Subjects, 1850(6) (2015), 1131-1139
https://doi.org/10.1016/j.bbagen.2015.02.003
RNA modulation of the human DNA methyltransferase 3A
Holz-Schietinger C, Reich NO. Nucleic Acids Res., 40(17) (2012), 8550-8557
https://doi.org/10.1093/nar/gks537
Mutations in DNA Methyltransferase (DNMT3A) Observed in Acute Myeloid Leukemia Patients Disrupt Processive Methylation
Holz-Schietinger C, Matje DM, Reich NO. J. Bio. Chem., 287(37) (2012), 30941-30951
https://doi.org/10.1074/jbc.M112.366625
Oligomerization of DNMT3A Controls the Mechanism of de Novo DNA Methylation
Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. J. Bio. Chem., 286(48) (2011), 41479-41488
https://doi.org/10.1074/jbc.M111.284687
Identification of a second DNA binding site in human DNA methyltransferase 3A by substrate inhibition and domain deletion
Purdy MM, Holz-Schietinger C, Reich NO. Arch. Biochem. Biophys., 498(1) (2010), 13-22
https://doi.org/10.1016/j.abb.2010.03.007
The Inherent Processivity of the Human de Novo Methyltransferase 3A (DNMT3A) Is Enhanced by DNMT3L
Holz-Schietinger C, Reich NO. J. Biol. Chem., 285(38) (2010), 29091-29100
https://doi.org/10.1074/jbc.M110.142513